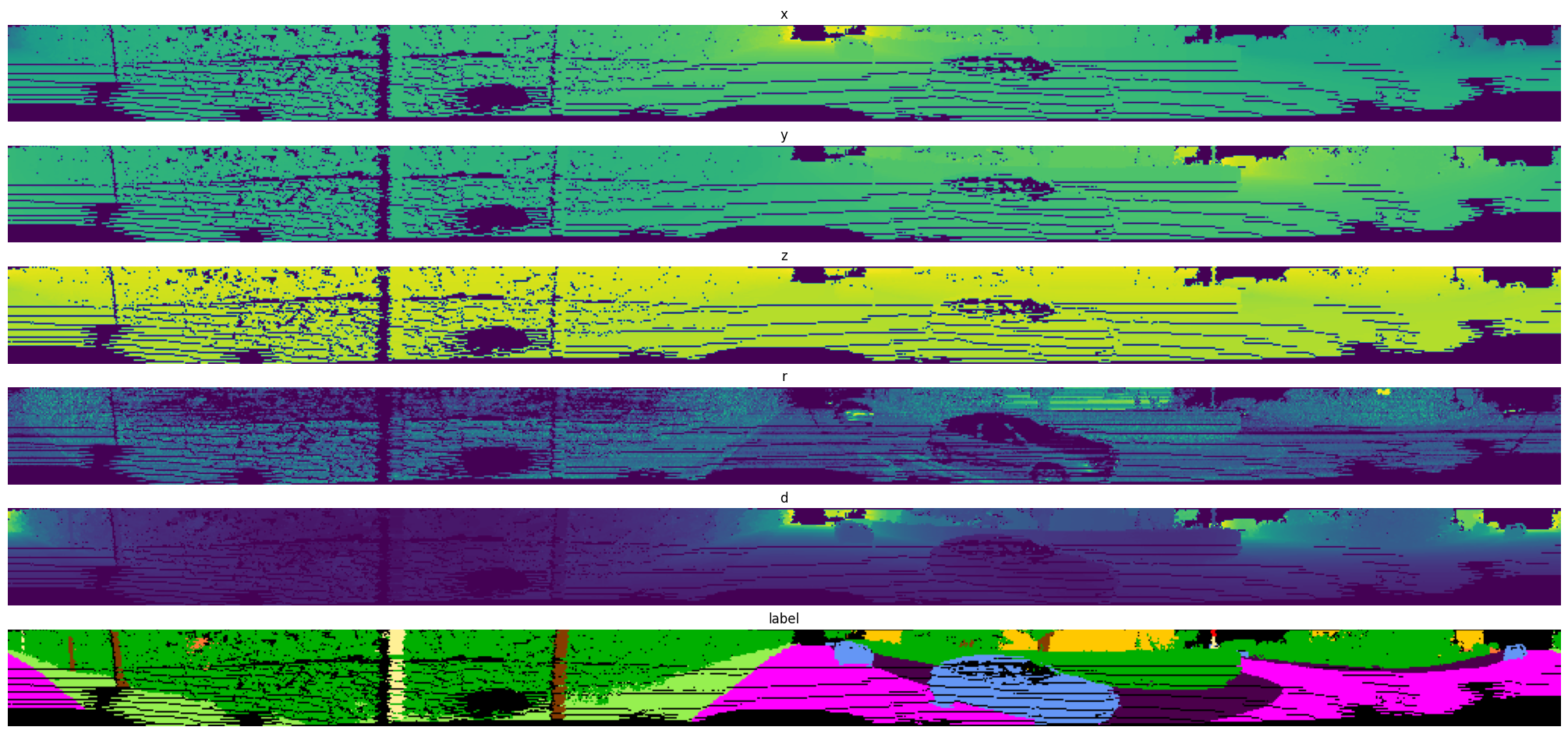
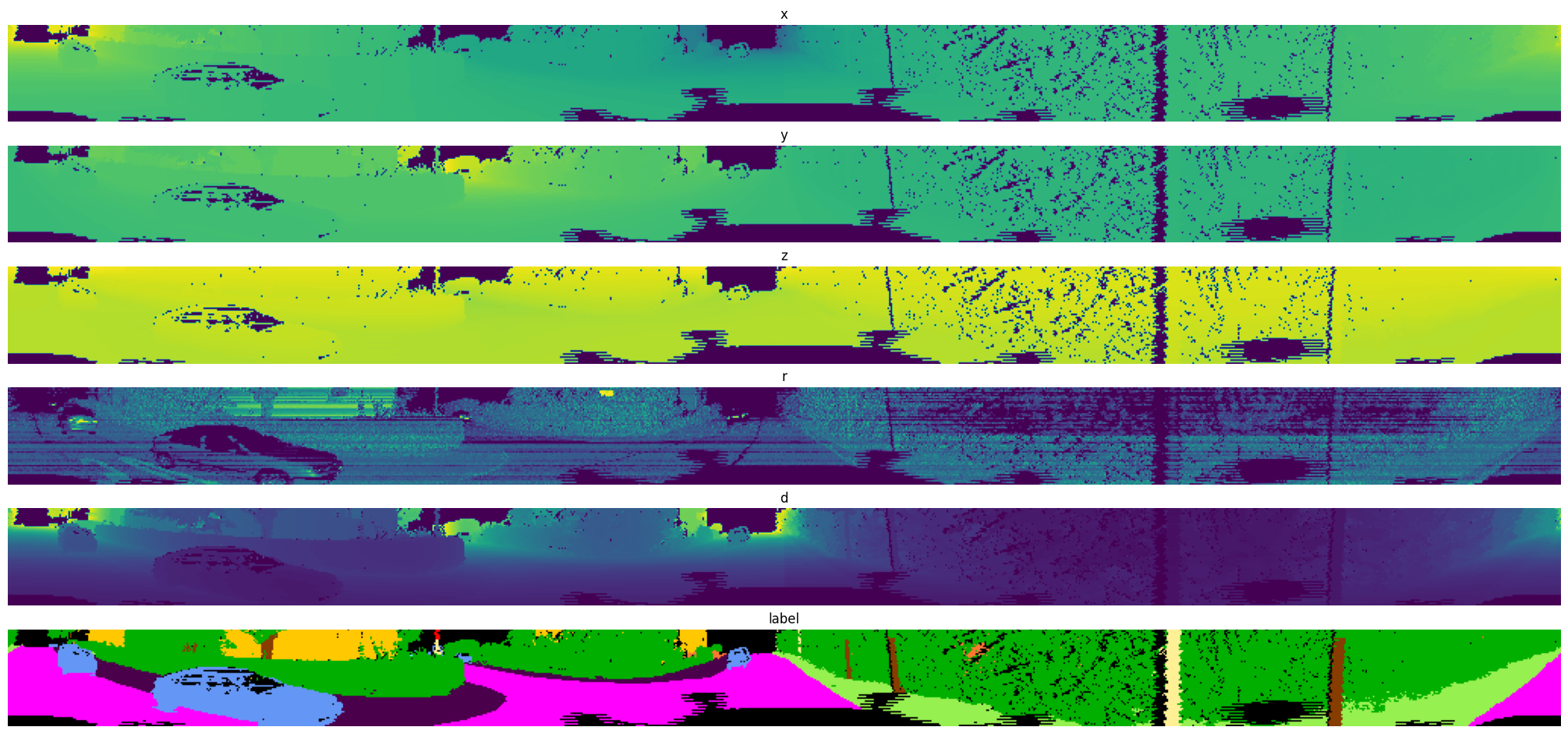
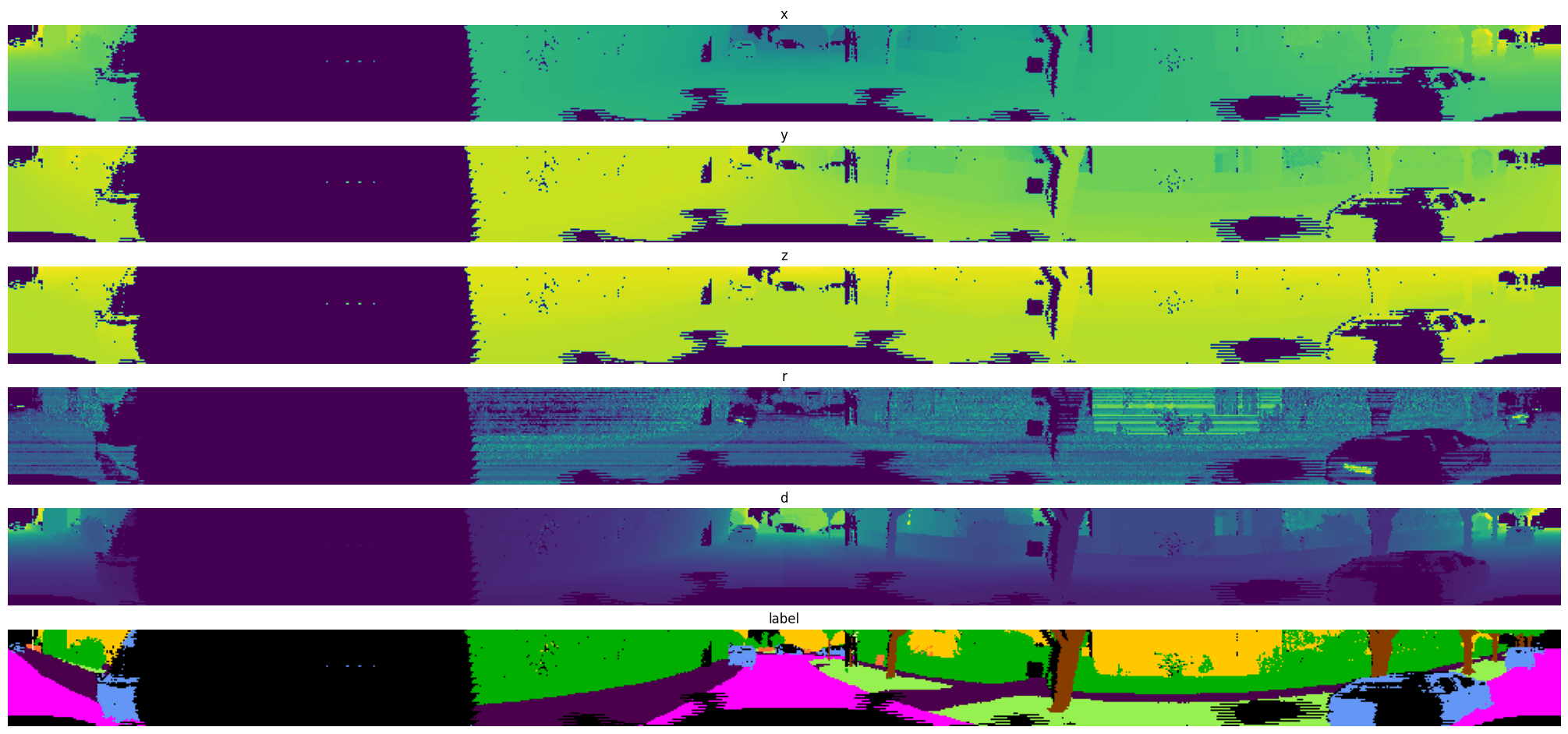
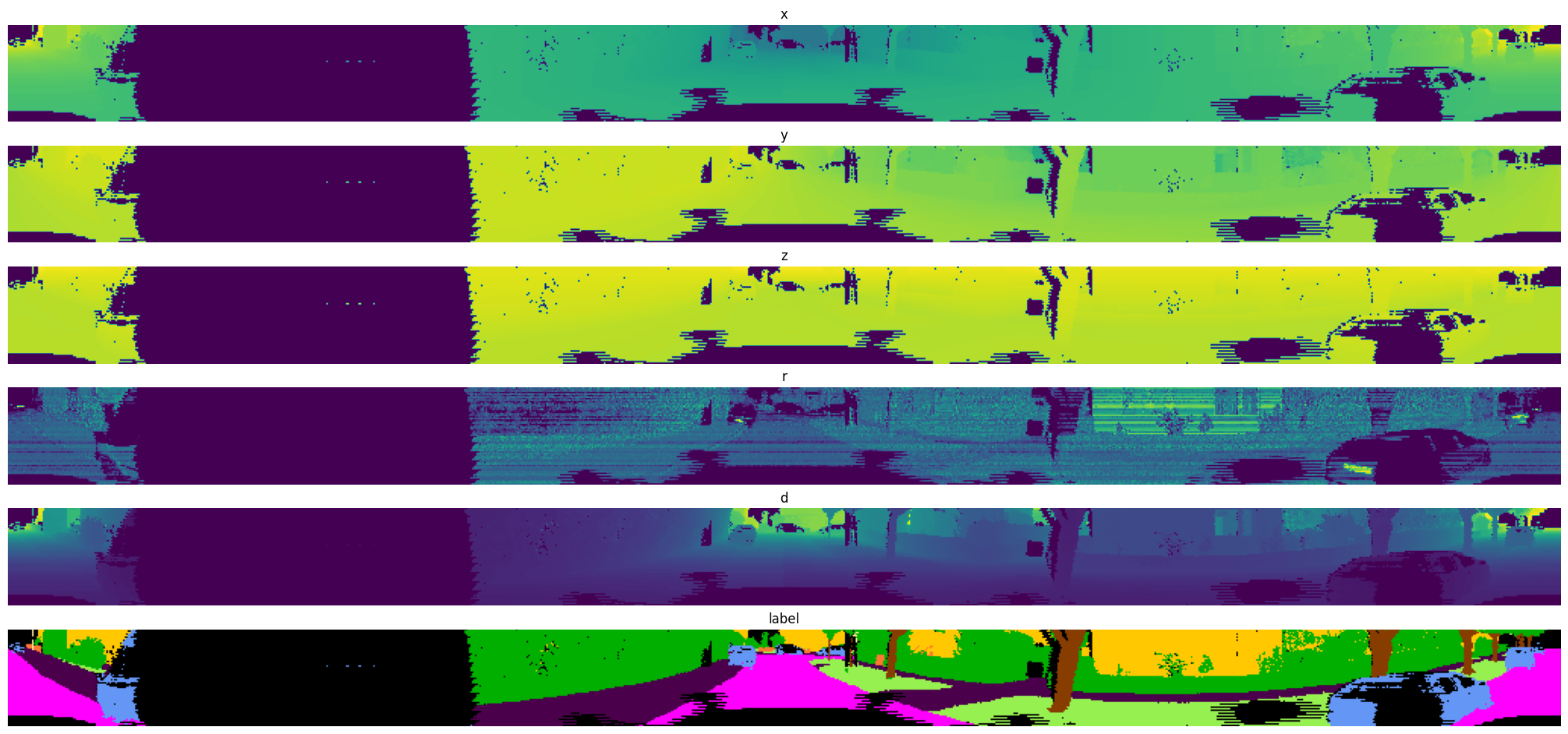
Exported source
class SemanticKITTIDataset(Dataset):
"Load the SemanticKITTI data in a pytorch Dataset object."
def __init__(self, data_path, split='train', transform=None, remapping_rules=None):
data_path = Path(data_path)
yaml_path = data_path/'semantic-kitti.yaml'
self.velodyne_path = data_path/'data_odometry_velodyne/dataset/sequences'
self.labels_path = data_path/'data_odometry_labels/dataset/sequences'
with open(yaml_path, 'r') as file:
metadata = yaml.safe_load(file)
if split != 'none':
sequences = metadata['split'][split]
velodyne_fns = []
for seq in sequences:
velodyne_fns += list(self.velodyne_path.rglob(f'*{seq:02}/velodyne/*.bin'))
self.frame_ids = [fn.stem for fn in velodyne_fns]
self.frame_sequences = [fn.parts[-3] for fn in velodyne_fns]
self.split = split
self.labels_dict = metadata['labels']
self.content = metadata['content']
max_key = sorted(self.content.keys())[-1]
self.content_np = np.zeros((max_key+1,), dtype=np.float32)
for k, v in self.content.items():
self.content_np[k] = v
self.learning_map = metadata['learning_map']
self.learning_map_np = np.zeros((max_key+1,), dtype=np.uint32)
for k, v in self.learning_map.items():
self.learning_map_np[k] = v
self.learning_map_inv = metadata['learning_map_inv']
self.learning_map_inv_np = np.zeros((len(self.learning_map_inv),), dtype=np.uint32)
content_sum_np = np.zeros_like(self.learning_map_inv_np, dtype=np.float32)
for k, v in self.learning_map_inv.items():
self.learning_map_inv_np[k] = v
content_sum_np[k] = self.content_np[self.learning_map_np == k].sum()
self.content_weights = 1./content_sum_np
self.color_map_bgr = metadata['color_map']
self.color_map_rgb_np = np.zeros((max_key+1, 3), dtype=np.float32)
for k, v in self.color_map_bgr.items():
self.color_map_rgb_np[k] = np.array(v[::-1], np.float32)
self.transform = transform
self.is_test = (split == 'test')
if remapping_rules is not None:
self.learning_remap(remapping_rules)
def learning_remap(self, remapping_rules):
new_map_np = np.zeros_like(self.learning_map_np, dtype=np.uint32)
max_key = sorted(remapping_rules.values())[-1]
new_map_inv_np = np.zeros((max_key+1,), dtype=np.uint32)
for k, v in remapping_rules.items():
new_map_np[self.learning_map_np == k] = v
if new_map_inv_np[v] == 0:
new_map_inv_np[v] = self.learning_map_inv_np[k]
new_content_sum_np = np.zeros_like(new_map_inv_np, dtype=np.float32)
for k in range(len(new_map_inv_np)):
new_content_sum_np[k] = self.content_np[new_map_np == k].sum()
self.learning_map_np = new_map_np
self.learning_map_inv_np = new_map_inv_np
self.content_weights = 1./new_content_sum_np
def set_transform(self, transform):
self.transform = transform
def __len__(self):
return len(self.frame_ids)
def __getitem__(self, idx):
assert self.split != 'none'
frame_id = self.frame_ids[idx]
frame_sequence = self.frame_sequences[idx]
frame_path = self.velodyne_path/frame_sequence/'velodyne'/(frame_id + '.bin')
with open(frame_path, 'rb') as f:
frame = np.fromfile(f, dtype=np.float32).reshape(-1, 4)
label = None
mask = None
if not self.is_test:
label_path = self.labels_path/frame_sequence/'labels'/(frame_id + '.label')
with open(label_path, 'rb') as f:
label = np.fromfile(f, dtype=np.uint32)
label = label & 0xFFFF
label = self.learning_map_np[label]
mask = label != 0 # see the field *learning_ignore* in the yaml file
item = {
'frame': frame,
'label': label,
'mask': mask
}
if self.transform:
item = self.transform(item)
return item